|
|
|
 |
Research in the group has focused on many problems of chemical synthesis, thermodynamic properties, and the structure/function of oligoribonucleotides and RNA. Apart from oligoribonucleotides and RNA containing natural nucleotides, we are interested in the studies of oligomers containing modified nucleotides. "RNA chemistry" in the group is not only a tool for the chemical synthesis of model oligoribonucleotides but also an approach for understanding the various behaviors and functions of RNA in nature. Recently, we have been working intensively on the chemical synthesis of locked nucleic acids (LNA), particularly the synthesis of LNA nucleotides containing natural and modified nucleobases. One of the major applications of modified oligoribonucleotides has been for constructing microarrays, which we are using to study the structure and interactions of RNA.
We have been involved in the study of the thermodynamic properties of nucleic acids since 1984, when we began collaborating with Professor Douglas H. Turner (University of Rochester, Rochester, USA). Our recent studies have focused on thermodynamic stability of LNA-2'-O-methylRNA/RNA chimeric duplexes containing modified nucleotides in the 2'-O-methylated strand. The LNA modified nucleotides include the full suite of naturally occurring bases (LNA G, C, A, U), as well as the non-natural bases: LNA-2,6-diaminopurine riboside, LNA-2-thiouridine, LNA-4-thiouridine and various 2'-O-methyl analogs of these compounds. We have determined the positional influence of single modified nucleotide substitutions on the thermodynamic stability of 2'-O-methylRNA/RNA duplexes. Based on thermodynamic data of 2'-O-methylRNA/RNA and LNA-2'-O-methylRNA/RNA duplexes, nearest neighbor rules for the stabilities of such chimeric duplexes were derived. Recently, we discovered that the addition of a 3' pyrene pseudonucleotide to an LNA-2'-O-methylRNA/RNA duplex universally enhances the thermodynamic stability of such duplexes. Interestingly, this effect seems to be independent of the type of base that is opposite of the pyrene pseudonucleotide. With these energy rules, it becomes possible to design finely-tuned modified oligonucleotides for numerous biological applications.
One of the most significant applications of our thermodynamic knowledge about these modified oligonucleotides has been demonstrated in the preparation of isoenergetic RNA microarrays. Using our energy rules we designed 853 isoenergetic pentanucleotides (out of 1024 possible 5-mers). The term isoenergetic is used, because the probes were designed in such a way that the thermodynamics of duplex formation across the set are roughly equal. Pentanucleotides were synthesized using the phosphoramidite approach incorporating our modified nucleosides. Currently, the library of isoenergetic probes allows for the printing of semi-universal microarrays (able to probe 853 out of 1024 possible fully complimentary binding sites). With this semi-universal coverage, it is possible to map the oligonucleotide accessibility of almost any folded RNA molecule. This binding profile is extremely sensitive to the target RNA structure and can be used to rapidly acquire data on the structure and interactions of the target of interest.
Our lab has applied isoenergetic microarray mapping to determine the secondary structures of a number of biologically interesting RNA molecules. Mapping was done to solve the structures of functionally important fragments from five recently sequenced R2 5'RNA retrotransposons from the silk moth family. We have also analyzed regulatory elements from Escherichia coli - DsrA RNA and OxyS RNA. In addition to the analysis of secondary structure we are also working on application of microarray mapping for the determination of interactions of both regulatory RNAs with RNA-binding Hfq protein. Recently, we expanded our research to use isoenergetic microarrays to scan disease associated RNAs (e.g. viral RNAs) for probes that bind strongly. Such strongly binding oligonucleotides may be used to inhibit the process of pathogenesis. As a model for those investigations we are using fragment X of the 3'UTR of Hepatitis C Virus.
We also are working on developing a method for the allele-specific digestion of pathogenic RNA based on an antisense oligonucleotides strategy. In our approach we are using gapmer type strongly modified antisense oligonucleotides. The differences in thermodynamic stability of complementary and single mismatched LNA-DNA/RNA duplexes are used to achieve specific hydrolysis at the point of the single nucleotide polymorphism (SNP) of a pathogenic RNA in the presence of ribonuclease H.
chemical synthesis of nucleosides, nucleotides and oligoribonucleotides containing modifications such as 2'-O-methylation, LNA backbones, and modified bases;
determination of the thermodynamic stability of RNA and DNA duplexes containing natural and modified nucleotides (modified 2'-O-methylnucleotides, LNA nucleotides, and modified LNA nucleotides);
construction of isoenergetic RNA microarrays based on short, modified pentanucleotide probes;
application of isoenergetic RNA microarrays (microarray mapping) to determine the structure of RNA and interactions with other biomolecules;
allelespecific hydrolysis of pathogenic RNA with modified antisense oligonucleotides.
Thermodynamic of LNA-2'-O-methylRNA/RNA duplexes containing modified nucleotides in 2'-O-methylated strand.
In collaboration with Douglas H. Turner, we studied the thermodynamic stability of 2'-O-methylRNA/RNA duplexes containing LNA-nucleotides and modified LNA-nucleotides. We found that the presence of LNA nucleotides enhance the thermodynamic stability (free energy, ∆Go37) of 2'-O-methylRNA/RNA duplexes by 1.4-2.4, 0.3-0.6 and 0.1-1.4 kcal/mol when placed at central, 5'- and 3'-terminal position within the duplex, respectively (see below). Moreover, substitution of LNA-adenosine by LNA-2,6-diaminopurine riboside additionally increases thermodynamic stability by ca. 1 kcal/mol. Recently, we also found that the placing pyrene pseudonucleotide at 3'-side of 2'-O-methylRNA strand of 2'-O-methylRNA/RNA duplexes enhances thermodynamic stability evenly by ca. 2.3 kcal/mol, no matter the nature of the nucleotide in the opposing RNA strand. Moreover, the influence of single mismatches on thermodynamic stability of 2'-O-methylRNA/RNA duplexes were also established and we found that the presence of single mismatch diminish thermodynamic stability of duplexes by 1.6-7.1 kcal/mol, depending on the type of mismatch. Based on collected data the thermodynamic parameters of nearest-neighbors for 2'-O-methylRNA/RNA and LNA-2'-O-methylRNA/RNA duplexes were calculated.
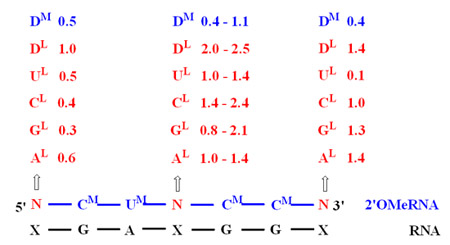
Schematic presentation of LNA-nucleotides influence on enhancement of the thermodynamic stability (∆Go37) of 2'-O-methylRNA/RNA duplexes (nucleotide X in RNA strand is complementary to LNA-nucleotide).
Developing a new type of microarray based on pentanucleotide isoenergetic probes (isoenergetic RNA microarrays).
A new method of RNA probing (microarray mapping) has been developed to study the structure and interactions of RNA. These microarrays are composed of penta/hexanucleotide probes, which are modified with various LNA-modified nucleotides such that the hybridization duplexes formed between oligonucleotide probes on the microarray and complementary fragments of target RNA are equally stable (as much as possible, forming an isoenergetic set). Additionally the modifications are designed to make the chimeric duplexes more favorable than unmodified duplexes. Some probes contain an additional LNA-guanosine at 3'-side, which additionally enhances the stability of duplexes by ca. 1.5 kcal/mole in a non-specific manner (unless it forms a C-G pair, which gives stronger enhancement). Only the presence of single stranded fragments of target RNA determines hybridization results due to the use of isoenergetic probes (in opposite to nonisoenergetic microarrays for which thermodynamic stability of hybridization duplexes is also key factor). This gives isoenergetic microarrays high structure sensitivity, but low sequence specificity. Presently, we have 853 penta/hexanucleotide probes, and all can form hybridization duplexes more favorable than -6 kcal/mol.
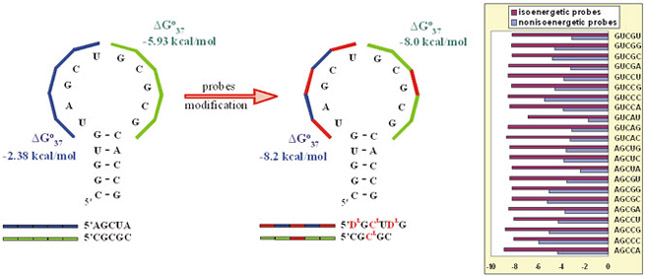
Schematic transformation of nonisoenegetic (unmodified) probes (left side) into isoenegetic (modified) probes (right side) with LNA-nucleotides and modified LNA-nucleotides. To the right is a comparison of binding free energies of nonisoenergetic (blue bars) and isoenergetic (purple bars) pentanucleotides.
Microarray mapping as new method to study structure of native RNAs and their interactions with other biomolecules.
Isoenergetic RNA microarrays offer a new, rapid method to study RNA structure, providing information on the location of single stranded fragments of targets. Results from microarray mapping can be used in structure prediction programs such as Mfold or RNA structure to increase the accuracy of predictions. The structure of several RNAs were studied including retrotransposon R2 5'RNAs from silk moths, that are known to possess important biological functionality. Microarray mapping was also applied to study regulatory RNA from Escherisia coli - DsrA RNA and OxyS RNA. Beside studies aimed at secondary structure we are also working on using the isoenergetic microarray method to gain insight into the interactions of both regulatory RNAs with Hfq protein and fragments of the regulated RNA.
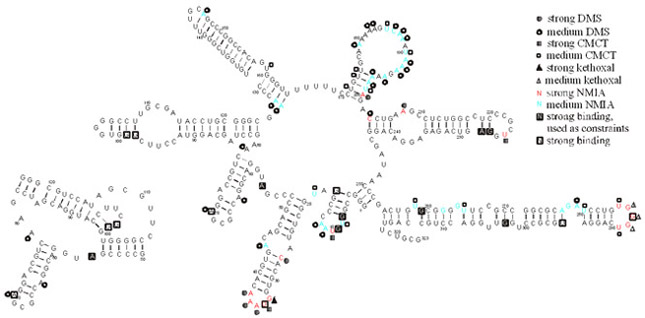
Prediction of secondary structure of R2 Bombyx mori 5' RNA from RNAstructure 4.4 using chemical mapping and microarray hybridization constraints or hybridization constraints alone at room temperature in buffer 200 mM NaCl, 5 mM MgCl2, 10 mM Tris-HCl, pH 8.0. On the left, fragment R2 5'RNA between 50 and 124 nucleotides which form pseudoknot structure (not predictable by RNAstructure program).
|
 |
|
|
